Do the COVID-19 injections contain epigenetic factors inducing cancer, autoimmunity, neurological disorders diabetes and more?
Is the spike protein also an epigenetic factor?
I just adore the following schematic diagram and I chose to post it here because soon, we might not be able to re-post even something as simple as a schematic from a website or a published journal article. (That’s why CAMPFIRE WIKI is so important, by the way.) This schematic is just so… comprehensive. The chromosome, the DNA, the histones, the histone tails, the methyl groups, the genes, the epigenetic factors, the health endpoints… sigh. It’s beautiful. I wonder how accurate it is?
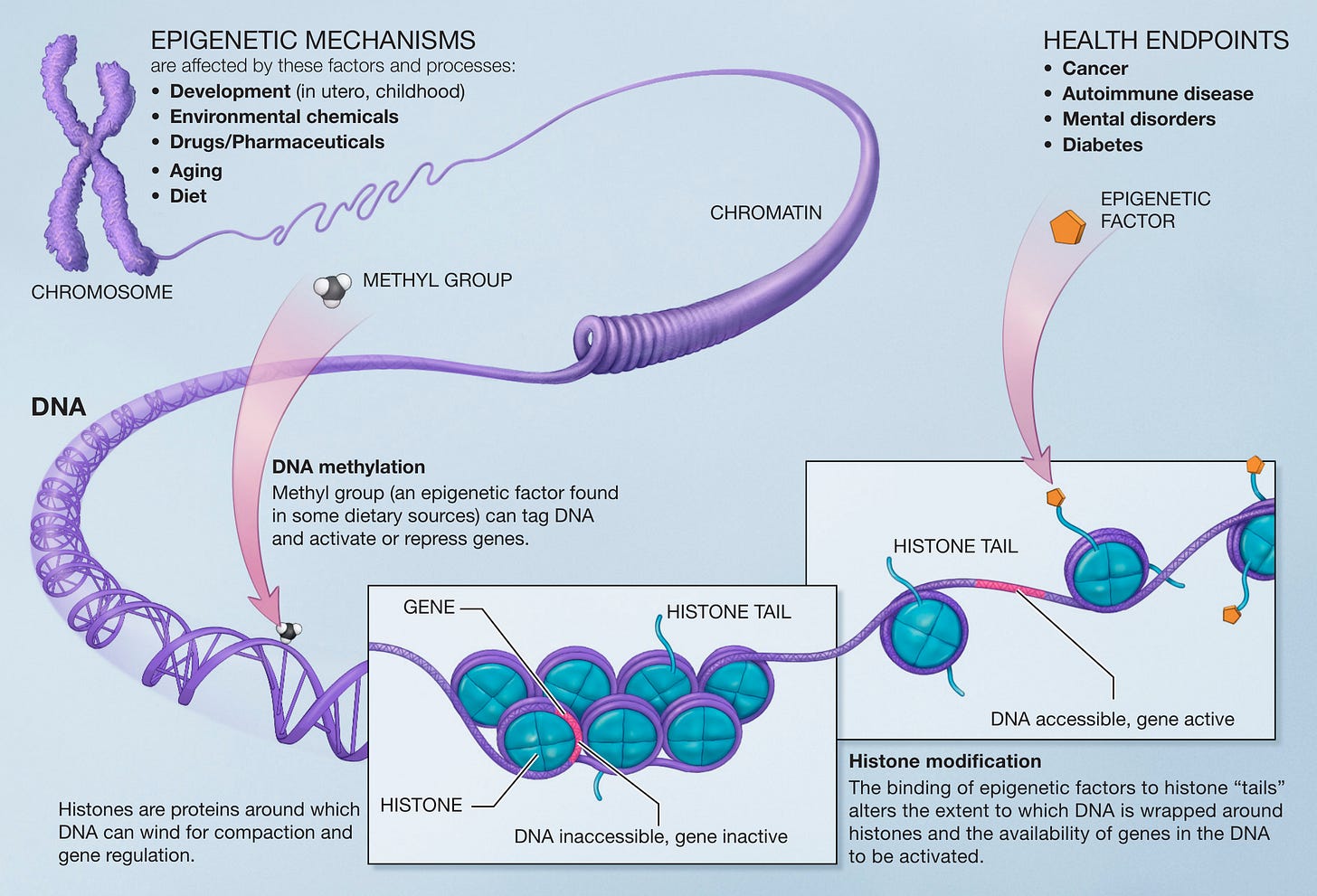
On epigenetics
The word epigenetics is made up of the prefix ‘epi’ from the Greek ἐπι, eti; meaning ‘over, outside of, around, or ‘in addition to’, and the suffix ‘genetics’ from the science of genes which comprises the traditional basis for inheritance.1 A gene is a chunk of coding material. DNA comprises a sequence of many genes. It also etymologically originates from the Greek γένος, génos; meaning generation or birth.2 It is interesting to me in these times of casual gene therapy use, that ‘gene therapy’ then etymologically necessarily implies the ‘curing’ of ‘generation or birth’. The word therapy comes from the Greek therapeia meaning “curing, healing, service done to the sick”.3 Now, this could be perfectly descriptive: to cure using genes. But, considering the data arising in these times surrounding declining fertility and birth rates in both females and males, it is worth paying attention to the specific effects of the COVID-19 shots, which are, in fact, mRNA based gene-therapy, on this facet of biological concerns.
Epigenetic changes involve changes in expression or activity of genes - NOT DNA (DeoxyriboNucleic Acid) - to alter cellular and physiological phenotypic traits resulting from external or environmental epigenetic factors. Epigenetics can also be used to describe any heritable phenotypic change and is part of normal development. It is very important to understand that the role of epigenetics in normal development is essential, and is precisely how and why we can have many different cell types, for example. Problems arise in terms of ‘disease’ (from Latin dis; ‘lack of’ and Old French aise; ‘well-being or comfort’4 5 implying a lack of well-being) in the context of impairment of this vital system that can arise with epigenetic modification.
Please watch the following video to get a good idea of what this gene expression and alteration and DNA stuff is all about, and why your ‘environment’ and microbiome are so vitally important to epigenetics and your health and well-being, in general.
On genomes in the context of epigenetics
All of the genetic material in your body comprises your genome and amazingly, each and every cell in your body contains all of the genetic material in your genome. By the way, your genome is yours and it is unique.6 Except in the case of say, twins. Twins have identical genomes, but they are different. Can you guess why? Amazingly, if you laid out your genome on the floor (don’t try to do this), it would measure approximately 2 meters long! That's like, almost as long as my Takayama noserider! So how does that all that DNA fit inside the nuclei of your little cells? It is by way of the magic of specialized proteins called histones.
Histones are octamers (they have 2 sets of identical 4 chunk units (H2A, H2B, H3 and H4) = octa (8)) that DNA is spooled around. Think of it like thread wound around a spool. You can wind a very long piece of string around a single spool, right? This is a space saving technique designed by nature. Each DNA/histone unit is called a nucleosome. Bunches of nucleosomes are called chromatin and you can imagine that being like a series of spools interconnected by one single long thread, like what is illustrated at the bottom of Figure 2. Chromatin is condensed further to make up chromosomes - those X-shaped thingies essential to our biology. Think of all those spools linked by that single thread bunched together to make up an X-shaped basket now filled with more closely spaced spools of thread. Those would be chromosomes.

As I alluded to before, the ability to modify gene expression (the effect of a gene being turned on or off via promoters) is why each cell type in our bodies have the ability to be different. Muscle cells versus liver cells, as illustrated in the above video, are completely different, and impose different functions based on which genes are turned on and off. The key to gene expression and activity is the organization of the nucleosome and the architecture of the chromatin. Each nucleosome has histone tails (originating from the histone), that can be capped to modify the tightness of the wrapping of the DNA around the histone. Think of the histone as a wooden spool, and the histone tails like shaved splinters that project outward. And think of the caps like little wax buds melted onto the ends of the splinters so you don’t get poked by them.
There are 2 primary types of epigenetic modifications: DNA methylation and histone modification.
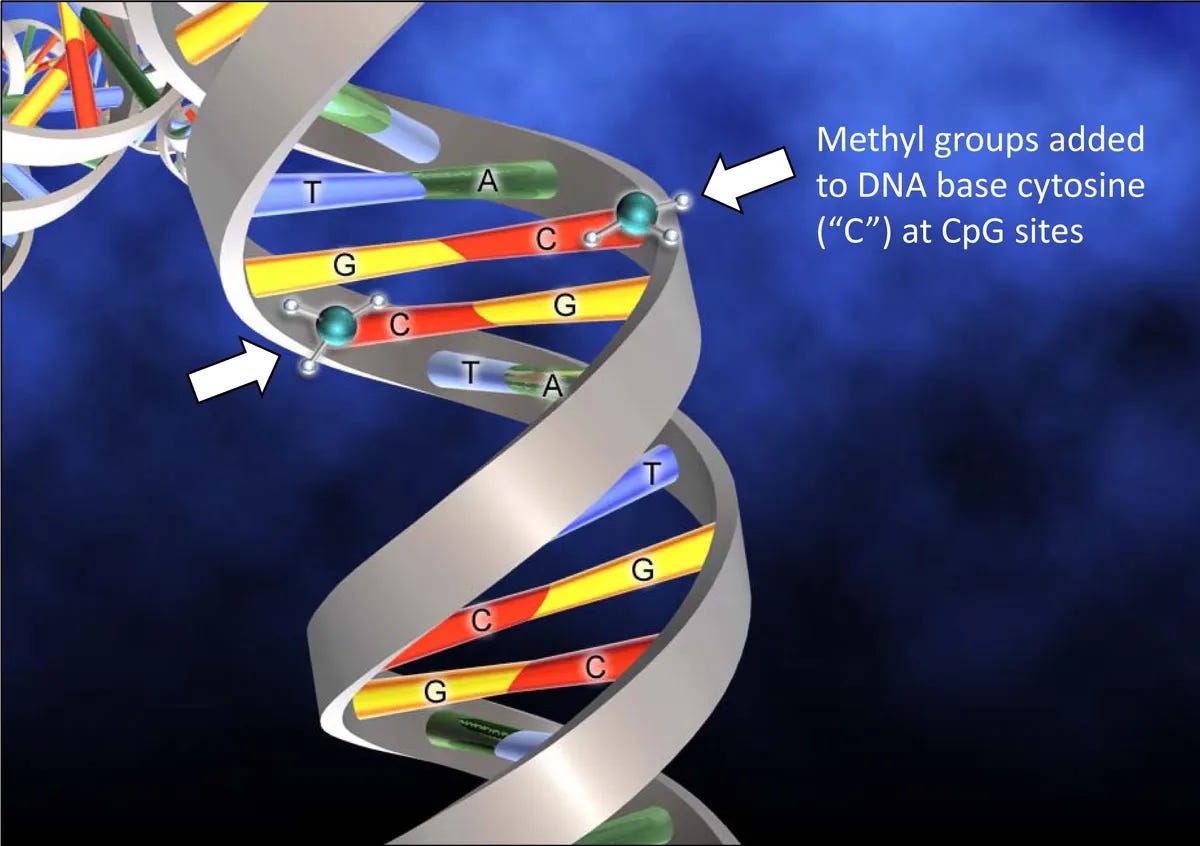
DNA methylation is when a methyl group is added directly to a cytosine residue that exists in a CpG sequence to induce gene silencing (shh, quiet gene). As all of my readers know by now, DNA is made up of four base nucleotides called cytosine (C), guanine (G), adenine (A) and thymine (T)). When a methyl group is attached to this cytosine site it is methylated, and when this methylation is in a promoter region, the gene associated with that C site will remain silent - it is not expressed. So methyl and acetyl groups linked to gene promoters are like being in the gene library where you have to be quiet. My jokes are getting really bad but trust me, they will get worse.
Histone modification, on the other hand, is when a methyl or acetyl group is added directly to specific residues on histone tails, subsequently modifying gene expression. Activation or repression of genes will depend on the residues modified, and the type of modification. Lysine acetylation correlates with transcriptional activation, for example.7
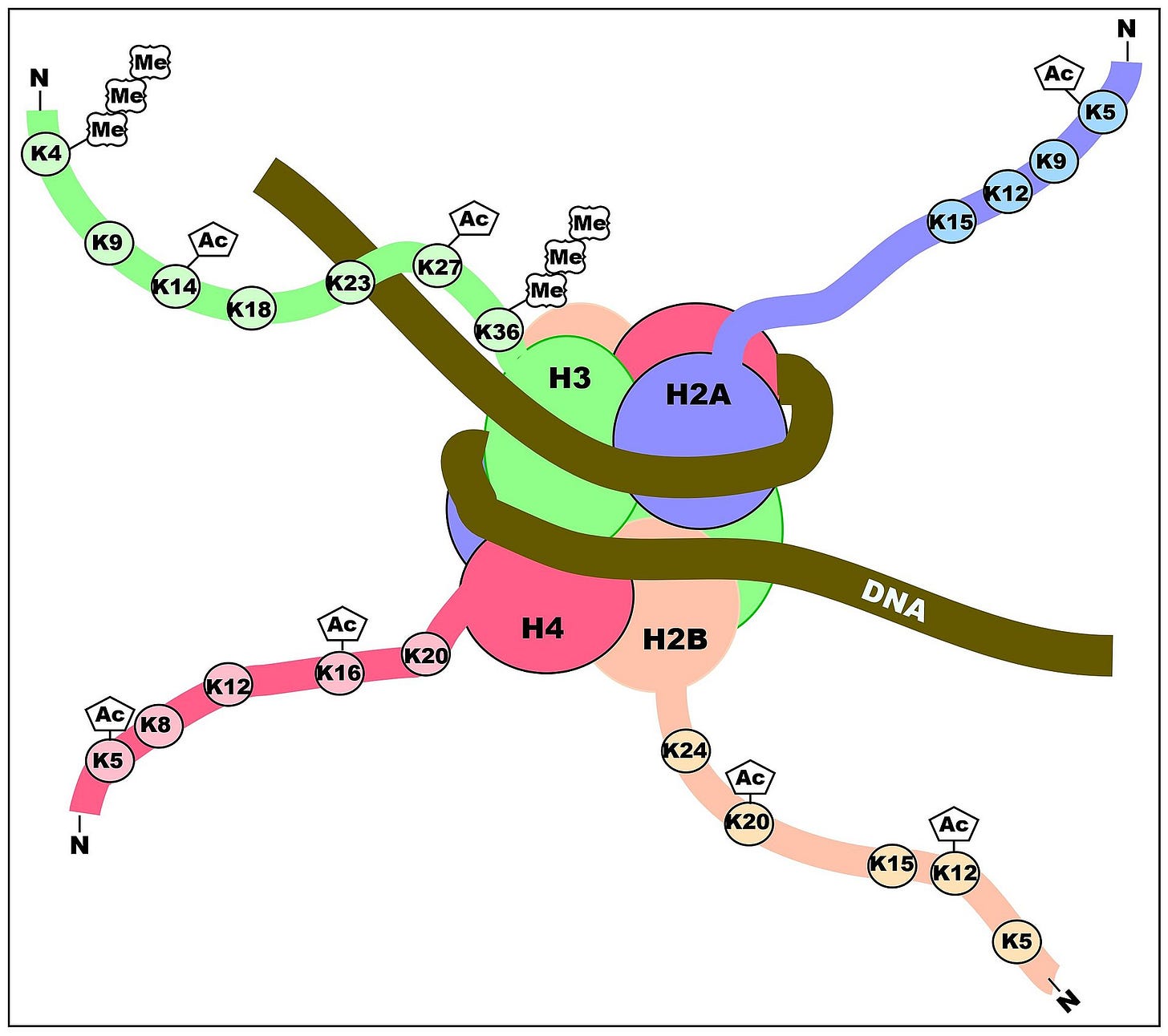
Histone modification via methylation and acetylation is basically the juice loosener. The juice is the histone, and the juice looseners are the methyl and acetyl groups. And yes, ‘p’ is for psycho. And the president’s neck is missing. Seriously though, using the ongoing spool and thread analogy, it is far more like maintaining or melting the wax buds off the ends of the wooden spool splinters so that, somehow, the thread is either tightly or loosely wound around the spools.
Methylation of histone tails that act to loosen up very tightly histone-wrapped DNA, can make the DNA accessible by other proteins that can subsequently read the genes therein for potential transcription. Note that histone tails can also undergo ubiquitylation, sumoylation and phosphorylation on specific residues.8 I have no idea what those are. Just kidding. I might explain phosphorylation later. Lysine acetylation correlates with transcriptional activation, for example.9 The methylation/demethylation patterns determine which genes are turned on and which are turned off and pretty much, determine the health state of the being.10
By the way, in addition to DNA methylation and histone modifications, epigenetic mechanisms involved in regulating gene expression and chromatin structure in human cells includes nucleosome positioning and histone variants and microRNAs (miRNA)s.11
On miRNAs
miRNAs are important in this context. To make a long story short, DNA has component parts called introns (do not code for proteins) and exons (code for proteins) that are both transcribable into messenger RNA. We all know what messenger RNA is by now, right? Introns, however, are never translated. They are removed from the messenger RNA via a process called splicing whereby the exons re-combine to form the translatable version of the messenger RNA. Well that is, when all things are right. Introns can be left behind (called intron retention) sometimes to cause problems like cancer.12

The introns, which our wise and dynamic science community used to refer to as junk DNA, comprise regulatory noncoding (not translated into proteins) RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and piwi RNA (piRNAs), which are all extremely important for gene regulation. They can all silence genes by regulation of production of proteins. Just because I can’t myself, if now was back then, no one would have been allowed to question the junk DNA hypothesis and thus, we probably wouldn’t know anything about miRNAs now. Something to think about.
miRNAs regulate gene expression by interfering with RNA or gene silencing. They come about via Drosha in the nucleus and Dicer in the cytoplasm. Each miRNA has a complementary messenger RNA that it can prevent from being translated to its protein. Kind of like a personal chastity belt if the miRNA is the belt and the messenger RNA is the person you want to dance to Barry White with.
On specific disease associations with epigenetic factors
Cancer is basically the epigenetic modification of the p53 tumor suppressor gene: the guardian of the genome. I love that: the guardian of the genome. I should actually be called p43.7, I think. I guess p53 is catchier.
The p53 protein not only enforces the stability of the genome by the prevention of genetic alterations in cells but also plays a role in regulating the epigenetic changes that can occur in cells.13
p53 suppresses tumor formation by regulating DNA repair. The impairment of this regulation can lead to cancer through loss of regulation of cell cycle. Its reintroduction into cancer cells, induces cell cycle arrest in the cancer cell. Expression levels, like everything in biology, is very important. Over expression of p53 can result in too much apoptosis (programmed cell death), and under expression can result in cancer.14 Epigenetic factors can affect p53 expression levels.15 16 17 18 19 They can also affect other DNA damage repair elements and their regulation.
A brilliantly frightening and frighteningly brilliant paper was published in October, 2021 in the journal Viruses entitled “SARS-CoV-2 Spike Impairs DNA Damage Repair and Inhibits V(D)J Recombination In Vitro”.20 When you read it, and you should read it, you will learn something very disturbing: the authors found that the spike protein was abundant in the nuclei of study cells. The nucleus is where most DNA repair occurs. They found that spike protein significantly suppressed both homologous recombination (HR) and non-homologous end joining (NHEJ) repair mechanisms. These are essential components to DNA repair and proper functioning in adaptive immunity (T and B cells). But even more scary, they found that the spike protein directly affects DNA repair in the nucleus by interfering with double-stranded DNA break (DSB) repair.
By the way, you might have noticed that there was an order to retract this work, and it was retracted. The claim written in the retraction request was strange and ambiguous, in my opinion, and it was made by the first author. The conclusion following ambiguous errors in methodology was that “conclusions related to vaccine safety are not validated and lacked experimental support”. Where have I heard this before? I will assume that the authors’ initial conclusions are the correct ones: I am suspicious of this retraction request. They write:
Here, by using an in vitro cell line, we report that the SARS–CoV–2 spike protein significantly inhibits DNA damage repair, which is required for effective V(D)J recombination in adaptive immunity. Mechanistically, we found that the spike protein localizes in the nucleus and inhibits DNA damage repair by impeding key DNA repair protein BRCA1 and 53BP1 recruitment to the damage site. Our findings reveal a potential molecular mechanism by which the spike protein might impede adaptive immunity and underscore the potential side effects of full-length spike-based vaccines.
Now, where have we heard of BRCA1 and 53BP1 before? Cancer context right? Breast cancer, right? BRCA1 is, in fact, a well-known breast cancer tumor suppressor gene and 53BP1 is a DNA damage checkpoint protein.21
What this means, is that evidence was published that proves that the spike protein is an epigenetic factor acting on DNA repair mechanisms to induce cancer. Period. And then it was retracted. Following peer review.
So either the original reviewers were all completely inept and missed this error that entirely debunks the conclusions of this work, or something squinky is going on here with the first author with regard to the retraction request.
This is where I put the brakes on this article. But I am going to methylate (I have gone full-on nerd) a few additional points because, as we all know, there’s more to disease than cancer.
On other disease associations with epigenetic factors
Bacterial infections are connected to epigenetics as well.22 23 I mean, literally, everything is on the list of epigenetic factors associated with disease. Everything. I am just going to list some of them with a single reference each.
Asthma,24 Hematologic malignancies,25 Alzheimer’s,26 Sperm motility,27 Chronic lymphocytic leukemia,28 Myocardial injury29 and cardiac hypertrophy,30 Autism,31 Frontal fibrosing alopecia,32 Diabetic Kidney Disease,33 Autoimmune conditions (in general - there are MANY),34 Cancer,35 COVID-1936 ... ruh roh
On solutions
Stop injecting yourselves with products with undisclosed ingredients that I, personally, am absolutely sure contain epigenetic factors causing all of the above, including cancer, by epigenetic modification.
There are a few reasons I provided background on miRNAs in this article. They regulate toll-like receptor (TLR) signaling pathways involved in inflammatory diseases.37 You can read about the connection between miRNAs, TLR-8 and severe COVID-19 here. Read Stephanie and Greg and Peter and Anthony’s paper to learn more about miRNA and SARS-nCoV-2/COVID-19 connections - they (3 specifically: miR-155, miR-148a and miR-590) are shipped around the body via exosomes to block type I interferon (IFN) responses and induce neuroinflammation, for example.38 I truly have not done this amazing paper remote justice in this summary, so please read it. It is long, but FULL of revealing information. Also, read everything Stephanie Seneff has ever written on glyphosate.
Another reason is that miRNAs can be used therapeutically, in theory, to aid in COVID-19 pathology.39
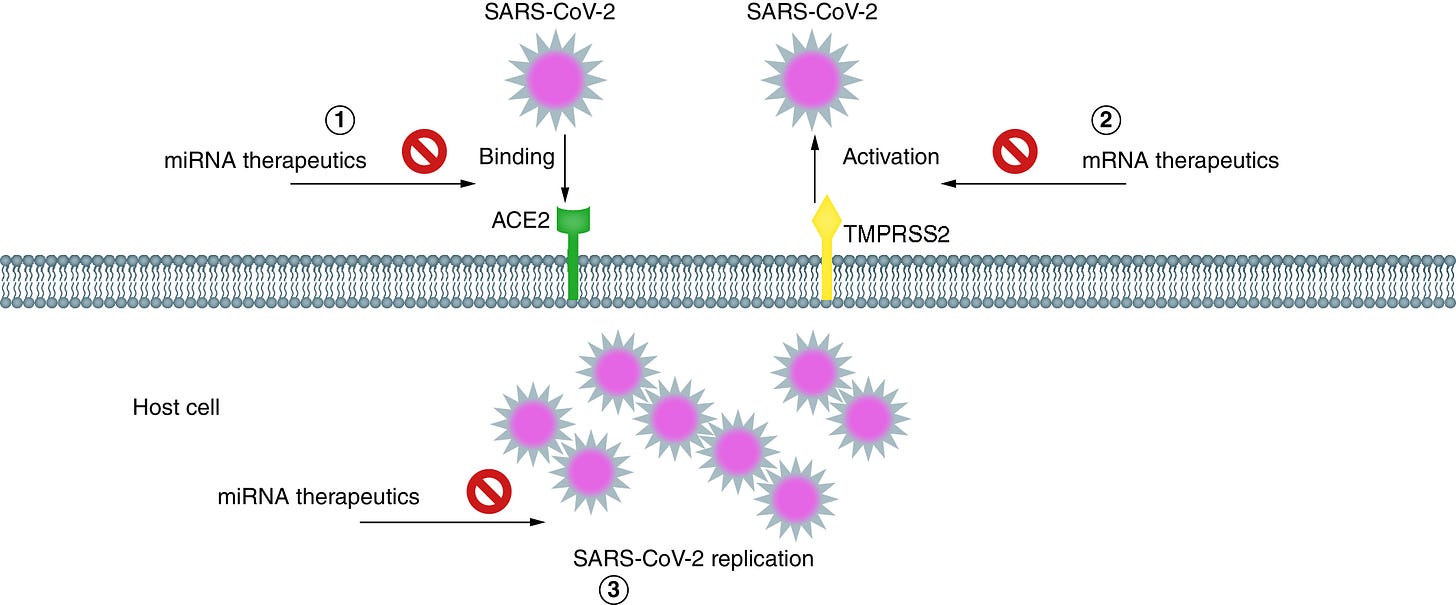
This isn’t over. Epigenetic modification covers just about everything disease-wise. It really does. Think about the past 100 years and think about differences between the East and the West. And read about inflammatory bowel disease and worms. Please head here for some more goodies with regard to scandals involving profit over well being.
Ultimately, we need balance and the chemical shit storm we are currently surviving through nowadays is not promoting this.
hashtagsickofbeingposioned
https://en.wikipedia.org/wiki/Epigenetics
https://en.wikipedia.org/wiki/Gene
https://www.etymonline.com/search?q=therapy
https://www.etymonline.com/word/ease?ref=etymonline_crossreference#etymonline_v_946
https://www.etymonline.com/word/dis-?ref=etymonline_crossreference
Varki A, Geschwind DH, Eichler EE. Explaining human uniqueness: genome interactions with environment, behaviour and culture. Nat Rev Genet. 2008 Oct;9(10):749-63. doi: 10.1038/nrg2428. PMID: 18802414; PMCID: PMC2756412.
Hebbes TR, et al. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402.
Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705.
Hebbes TR, et al. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402.
Neri F, Incarnato D, Oliviero S. DNA methylation and demethylation dynamics. Oncotarget. 2015 Oct 27;6(33):34049-50. doi: 10.18632/oncotarget.6039. PMID: 26461852; PMCID: PMC4741426.
Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010 Jan;31(1):27-36. doi: 10.1093/carcin/bgp220. Epub 2009 Sep 13. PMID: 19752007; PMCID: PMC2802667.
https://www.zmescience.com/medicine/genetic/intron-retention-cancer-25012016/
Arnold J. Levine and Shelley L. Berger. The interplay between epigenetic changes and the p53 protein in stem cells. GENES & DEVELOPMENT 31:1195–1201.
Lakin, N., Jackson, S. Regulation of p53 in response to DNA damage. Oncogene 18, 7644–7655 (1999). https://doi.org/10.1038/sj.onc.1203015.
Tomicic MT, Dawood M, Efferth T. Epigenetic Alterations Upstream and Downstream of p53 Signaling in Colorectal Carcinoma. Cancers (Basel). 2021;13(16):4072. Published 2021 Aug 13. doi:10.3390/cancers13164072.
Levine AJ, Berger SL. The interplay between epigenetic changes and the p53 protein in stem cells. Genes Dev. 2017;31(12):1195-1201. doi:10.1101/gad.298984.117.
Saldaña-Meyer R, Recillas-Targa F. Transcriptional and epigenetic regulation of the p53 tumor suppressor gene. Epigenetics. 2011;6(9):1068-1077. doi:10.4161/epi.6.9.16683.
Krepulat F, Löhler J, Heinlein C, Hermannstädter A, Tolstonog GV, Deppert W. Epigenetic mechanisms affect mutant p53 transgene expression in WAP-mutp53 transgenic mice. Oncogene. 2005;24(29):4645-4659. doi:10.1038/sj.onc.1208557.
Ward A, Hudson JW. p53-Dependent and cell specific epigenetic regulation of the polo-like kinases under oxidative stress. PLoS One. 2014 Jan 31;9(1):e87918. doi: 10.1371/journal.pone.0087918. PMID: 24498222; PMCID: PMC3909268.
Jiang H, Mei YF. SARS-CoV-2 Spike Impairs DNA Damage Repair and Inhibits V(D)J Recombination In Vitro [retracted in: Viruses. 2022 May 10;14(5):]. Viruses. 2021;13(10):2056. Published 2021 Oct 13. doi:10.3390/v13102056.
Daley JM, Sung P. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol Cell Biol. 2014 Apr;34(8):1380-8. doi: 10.1128/MCB.01639-13. Epub 2014 Jan 27. PMID: 24469398; PMCID: PMC3993578.
Bierne H, Hamon M, Cossart P. Epigenetics and bacterial infections. Cold Spring Harb Perspect Med. 2012 Dec 1;2(12):a010272. doi: 10.1101/cshperspect.a010272. PMID: 23209181; PMCID: PMC3543073.
Wang J, Liu Z, Xu Y, et al. Enterobacterial LPS-inducible LINC00152 is regulated by histone lactylation and promotes cancer cells invasion and migration. Front Cell Infect Microbiol. 2022;12:913815. Published 2022 Jul 25. doi:10.3389/fcimb.2022.913815.
Lavoie ME, Meloche J, Boucher-Lafleur AM, et al. Longitudinal follow-up of the asthma status in a French-Canadian cohort. Sci Rep. 2022;12(1):13789. Published 2022 Aug 13. doi:10.1038/s41598-022-17959-6.
Hofmann WK, Trumpp A, Müller-Tidow C. Therapy Resistance Mechanisms in Hematological Malignancies [published online ahead of print, 2022 Aug 13]. Int J Cancer. 2022;10.1002/ijc.34243. doi:10.1002/ijc.34243.
Singh AK, Neo SH, Liwang C, et al. Glucose derived carbon nanosphere (CSP) conjugated TTK21, an activator of the histone acetyltransferases CBP/p300, ameliorates amyloid-beta 1-42 induced deficits in plasticity and associativity in hippocampal CA1 pyramidal neurons [published online ahead of print, 2022 Aug 12]. Aging Cell. 2022;e13675. doi:10.1111/acel.13675.
Guo H, Shen X, Hu H, et al. Alteration of RNA modification signature in human sperm correlates with sperm motility [published online ahead of print, 2022 Aug 12]. Mol Hum Reprod. 2022;gaac031. doi:10.1093/molehr/gaac031.
Bruch PM, Giles HA, Kolb C, et al. Drug-microenvironment perturbations reveal resistance mechanisms and prognostic subgroups in CLL. Mol Syst Biol. 2022;18(8):e10855. doi:10.15252/msb.202110855.
He L, Wang Y, Luo J. Epigenetic modification mechanism of histone demethylase KDM1A in regulating cardiomyocyte apoptosis after myocardial ischemia-reperfusion injury. PeerJ. 2022;10:e13823. Published 2022 Aug 5. doi:10.7717/peerj.13823.
Han Y, Nie J, Wang DW, Ni L. Mechanism of histone deacetylases in cardiac hypertrophy and its therapeutic inhibitors. Front Cardiovasc Med. 2022;9:931475. Published 2022 Jul 26. doi:10.3389/fcvm.2022.931475.
Shirinian M, Chen C, Uchida S, Jadavji NM. Editorial: The role of epigenetics in neuropsychiatric disorders. Front Mol Neurosci. 2022;15:985023. Published 2022 Jul 25. doi:10.3389/fnmol.2022.985023.
Miao YJ, Jing J, Du XF, Mao MQ, Yang XS, Lv ZF. Frontal fibrosing alopecia: A review of disease pathogenesis. Front Med (Lausanne). 2022;9:911944. Published 2022 Jul 25. doi:10.3389/fmed.2022.911944.
Rico-Fontalvo J, Aroca G, Cabrales J, et al. Molecular Mechanisms of Diabetic Kidney Disease. Int J Mol Sci. 2022;23(15):8668. Published 2022 Aug 4. doi:10.3390/ijms23158668.
Mazzone, R., Zwergel, C., Artico, M. et al. The emerging role of epigenetics in human autoimmune disorders. Clin Epigenet 11, 34 (2019). https://doi.org/10.1186/s13148-019-0632-2.
Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010 Jan;31(1):27-36. doi: 10.1093/carcin/bgp220. Epub 2009 Sep 13. PMID: 19752007; PMCID: PMC2802667.
Delshad M, Sanaei MJ, Pourbagheri-Sigaroodi A, Bashash D. Host genetic diversity and genetic variations of SARS-CoV-2 in COVID-19 pathogenesis and the effectiveness of vaccination [published online ahead of print, 2022 Aug 8]. Int Immunopharmacol. 2022;111:109128.
Arenas-Padilla M, Mata-Haro V. Regulation of TLR signaling pathways by microRNAs: implications in inflammatory diseases. Cent Eur J Immunol. 2018;43(4):482-489. doi: 10.5114/ceji.2018.81351. Epub 2018 Dec 31. PMID: 30799997; PMCID: PMC6384427.
Seneff S, Nigh G, Kyriakopoulos AM, McCullough PA. Innate immune suppression by SARS-CoV-2 mRNA vaccinations: The role of G-quadruplexes, exosomes, and MicroRNAs. Food Chem Toxicol. 2022;164:113008. doi:10.1016/j.fct.2022.113008.
Fani M, Zandi M, Ebrahimi S, Soltani S, Abbasi S. The role of miRNAs in COVID-19 disease. Future Virol. 2021 Mar:10.2217/fvl-2020-0389. doi: 10.2217/fvl-2020-0389. Epub 2021 Mar 24. PMCID: PMC7989380.




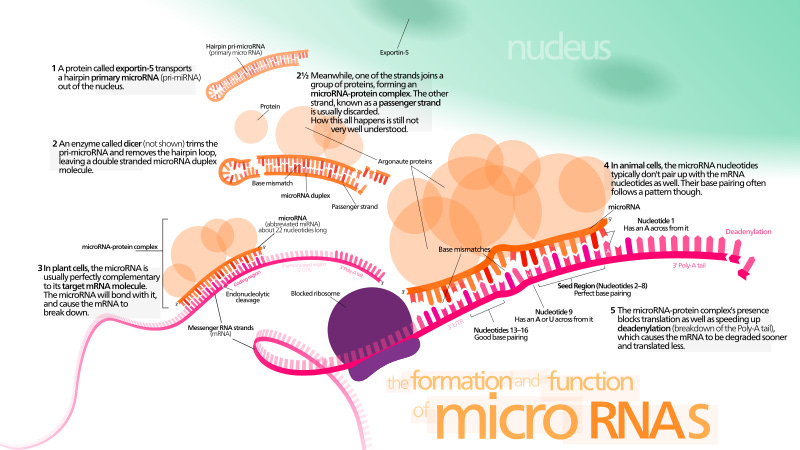
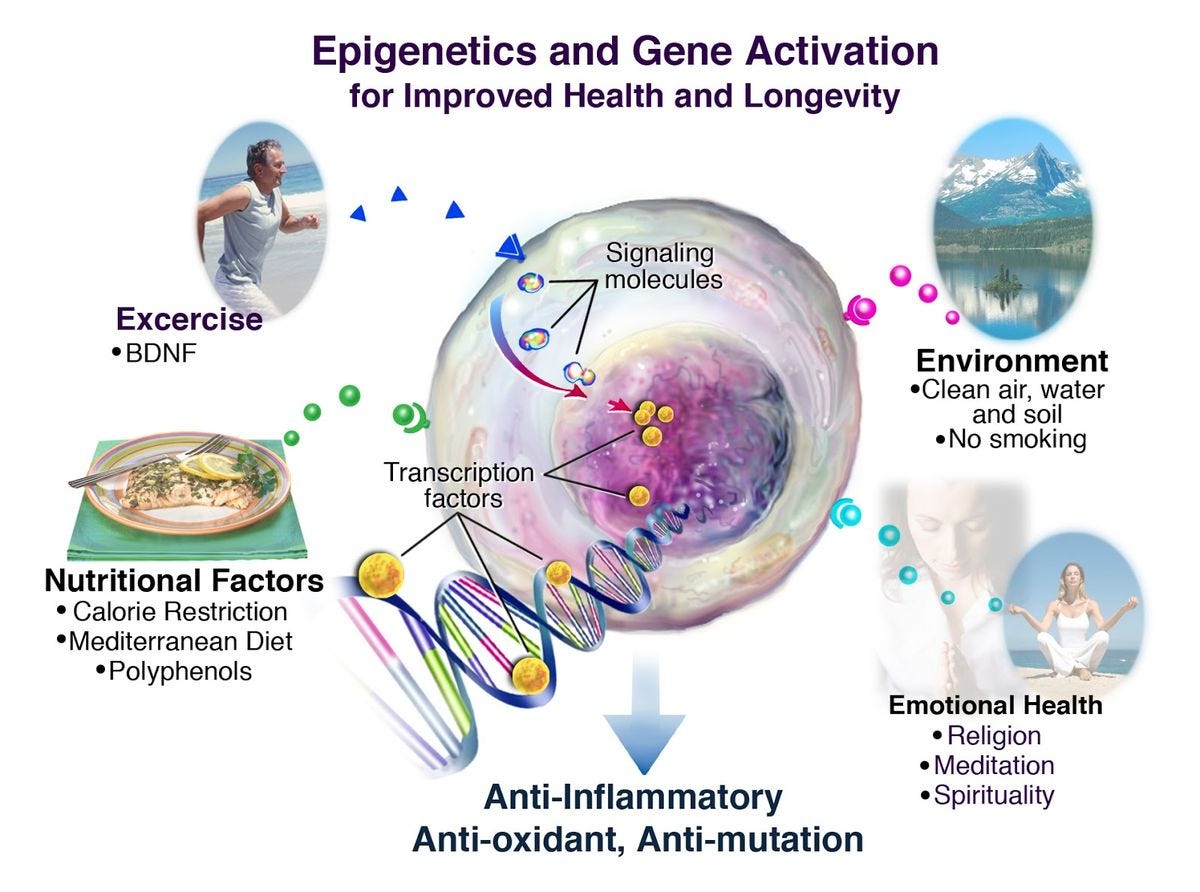
Brilliant. Thank you.
Why "because soon, we might not be able to re-post even something as simple as a schematic from a website or a published journal article." ? Sorry, under a rock ...